

CAPITALISE are adopting open data approaches
During the first reporting period, IPK, SSSA, and UP, in collaborations with the rest of the consortium, contributed to developing a research data management plan (RDMP) that provides a common framework for storing, accessing, and processing the diverse data sets generated in CAPITALISE, namely: sequencing data, phenotyping data metabolic profiling data physiological data phenological and yield data and socioeconomic data.
FAIR data principles are at the core of the CAPITALISE RDMP, and are realized by: (i) consortium agreement to use standardized formats and ontological descriptors, (ii) rendering data openly accessible via DOIs, (iii) employing consistent descriptors of genetic material, and (iv) conducting analyses based on open software applications or code that is made fully available to ensure reproducibility.
Metadata templates design at IPK
IPK have been working on generating metadata templates following the standardized format using MIAPPE (Krajewski et al. 2015) to provide all partners with a suitable and efficient way to store and handle data.
The aim of the templates is FAIR data handling (Wilkinson et al. 2016) and data publishing to PGP repositories such as e!DAL (Arend et al. 2014). The initial MIAPPE-compliant templates were iteratively customized to the needs of the partners of the consortium. The metadata templates are now divided into datasheets with static and dynamic attributes. A static datasheet includes all the static MIAPPE attributes throughout the project from the investigation, study, biological material, person, and observation unit sections. Dynamic attributes additionally allow users to store and archive information about changes of experimental nature or of certain events throughout the experiments.
An example of data capturing in a standardized way using MIAPPE compliant datasheets can be seen below, where unstructured raw datasets (upper sheets) received from partners and were transformed into structured Event datasheet (lower sheet).
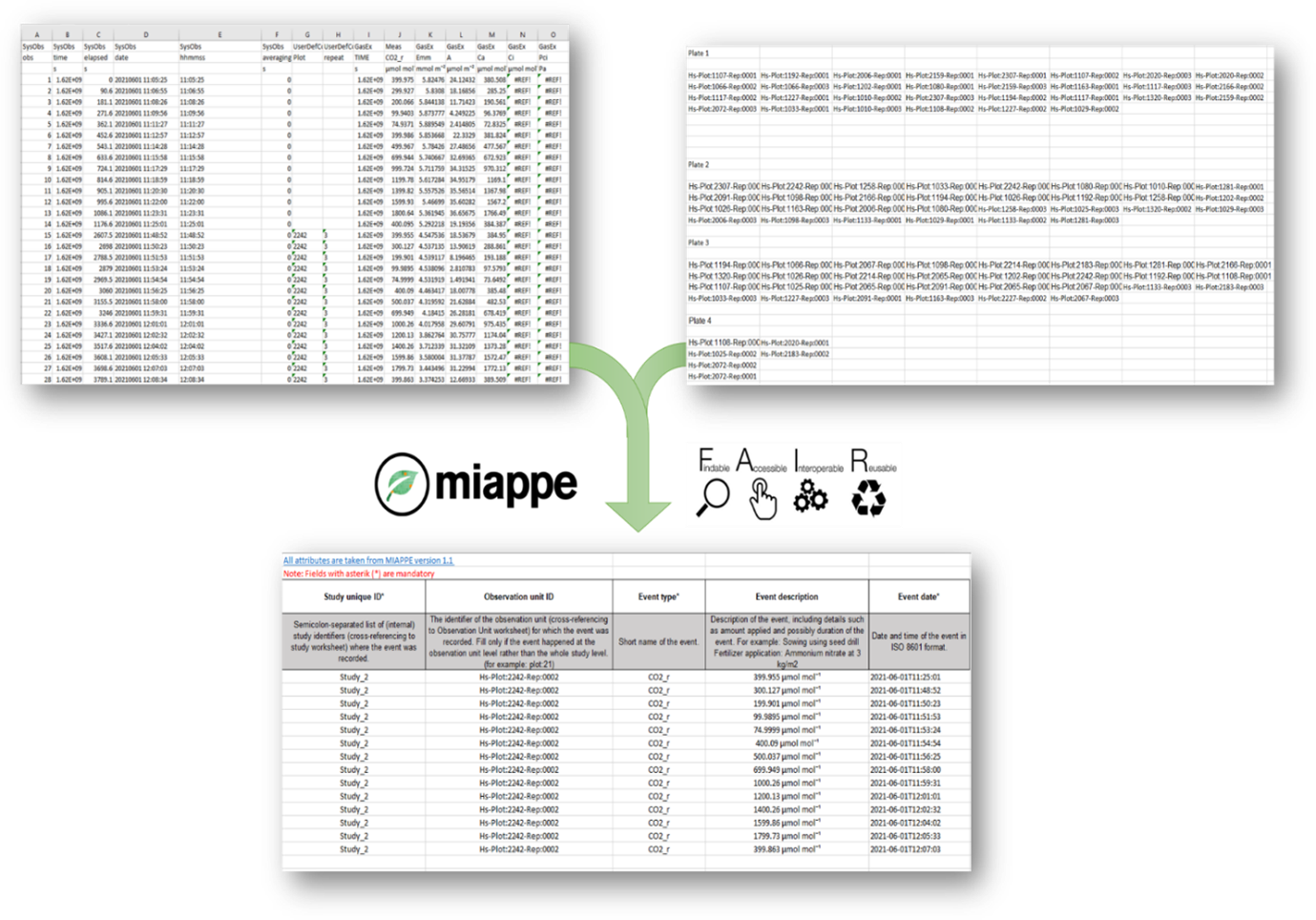
Transformation of unstructured data to MIAPPE compliant datasheets
References
- Arend D, Lange M, Chen J, et al (2014) e!DAL – a framework to store, share and publish research data. BMC Bioinformatics 15:214.
- Krajewski P, Chen D, Ćwiek H, et al (2015) Towards recommendations for metadata and data handling in plant phenotyping. J Exp Bot 66:5417–5427.
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al (2016) The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3:160018.


